SAMI Data Reduction Cookbook¶
Overview
This cookbook presents the typical procedure for the data reduction with SAMI (SOAR imager) using the PySOAR package.
Date: May 10, 2012 Author: Luciano Fraga email: lfraga@ctio.noao.edu
The document is intended for astronomers who want to reduce SAMI data using the PySOAR package. PySOAR is running only on soarpd1 computer. In order to use it, you must follow the steps outline in this cookbook: logging on soarpd1, transfer the data to soarpd1 and run the pipepline.
Logging On¶
Logging on from the Remote Observing Center in La Serena or Cerro Pachon.
Type the following command from a terminal on the GNU/Linux computer:
vncviewer -Shared soarpd1.ctio.noao.edu:9 &
or:
vncviewer -Shared 139.229.13.231:9 &
Note
userid is observer and vnc’s password is provided to you by me (Luciano Fraga)
From outside the CTIO network (Remote Observation).
To observe remotely, you will need a VPN connection
Note
please, contact your scientific staff for VPN passwords and usernames.
When you have successfully connected into soarpd1:9 you will see a desktop like that:

Transfering the data¶
The SAMI data is storage on soarhrc. To transfer the data from soarhrc to soarpd1 you must either click on the top-left icon:

or type in a terminal windown the following command:
data_transfer.py
The following window will appear:
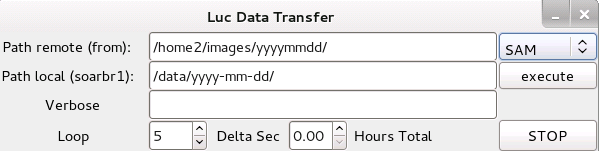
by (e.g.):
/home/observer/data/2012-03-01
Running the pipeline scripts¶
Go to the data directory (e.g. cd /home/observer/data/2012-03-01).
Suggestion: create a RED directory and copy all your data to it. Go to the RED directory.
Important! The calibration frames (biases and flat-fields) and the science frames must be in the same directory. If you already have the master bias and master flats, just copy both to the same directory.
Run the pipeline with the command line:
samipipe.py
- When the script is finished you are going to reduced files with the following nomenclature:
- The script will create lists of images as bellow:
- 0SAMIList_Zero1x1: List of biases with binning 1x1
- 1SAMIList_Flat1x1B: List of dome flat-fields with binning 1x1 and filter B.
- 2SAMIList_SFlat1x1B: List of sky flat-fields with binning 1x1 and filter B.
- 3SAMIList_Dark1x1: List of dark frames with binning 1x1.
- 4SAMIList_OBJ1x1B: List of science images with binning 1x1 and filter B.
- For a raw science frame like image.001.fits, the reduced frame will be like mzfimage.001.fits. The prefix z means zero subtract, f means flat-field divided and m means that the file has been converted from multi-extend fits to single fits.
Step-by-step Data Reduction¶
TODO: Incomplete!
samizerocombine: master bias
samiflatcombine: master flat-field (normalized,bias subtracted) for each filter
samireduce: apply to the science object
samimosaic: merge the ccds

