

The measurement of concentricities has to be made off telescope probably at the Coude Room. The minimal setup required to measure the concentricities includes, besides Hydra, a video monitor, an ICCD controller, a video cursor, a light source and the Hydra Laptop (or the Hydra PC). The video monitor plus the ICCD controller display the video signal from the gripper camera. The light source is used to illuminate the fibers from behind. And the Laptop controls the instrument. When illuminating the fibers environmental light has shown to be sufficient for the small and large fibers. To illuminate the FOPS it's not necessary to remove the FOPS camera. Removing the filter in front of the camera will allow you to introduce enough light through the space left by the filter.
If you are using the Hydra Laptop to control the instrument during this procedure you better be sure that the "CurrentLocations" and "statusFile" are not different from the ones in ctioa6 (Hydra PC). Copying this files from ctioa6 to the laptop will ensures that the software knows exactly where the fibers are. For example if the Ethernet connection is available, typing the following commands in a shell window will do it for you. Don't forget to login as "hydra".
% cd Hydra/data/status
% rcp ctioa6:/home/hercules/Hydra/data/status/CurrentLocations .
% rcp ctioa6:/home/hercules/Hydra/data/statuc/statusFile .
Now you can start "hydractio" or "hydractio-comm" if you don't want to communicate with the TCS and FOPS Guider systems (this will be often the case).
The canonical procedure is first to put the fibers in a circle. There is a script that can do this job for you. For example, if you are measuring the concentricities for the small fibers type at the CLI/Script Tool command line:
Fiborg3.2.0> execfile configsmallcircle
After it finishes you can execute the script to actually measure the offset between the button position and the fiber position for every fiber. Don't forget to check that your light source is illuminating the fibers evenly before executing the script. What the script does is to go fiber after fiber grabbing its current position, allowing you to correct the position by offsetting the gripper and grabbing the new position. The idea is to use a video reference (video cursor) to center, by moving the gripper, the light beam coming from the fiber and use the offset between the original (button position) and final position (fiber position) to calculate the concentricity correction for that particular fiber. You should repeat the process a minimum of three times (five is a good number of repetitions), that is parking all the fibers, putting them back in a circle and grabbing the corrections. Make sure you also do FOPS concentricities measurements interleaved with the small or large concentricities measurements. The FOPS serve as the zero point correction. It is recommended to do all the fibers (small, large and FOPS) with the same TV setup just to ensure that they are all on the same zero point. All the recorded positions are kept in the log file of the current session.
For example, if you want to record concentricity data for the small fibers you should type at the CLI prompt:
Fiborg3.2.0> execfile meassmall
The gripper will move to the first small fiber and will wait for you to move the gripper. You can use the arrow buttons in the "Field Display Tool" window or the keyboard by typing "h, j, k, l" (make sure the mouse cursor is on top of the "Field Display Tool" window) to offset the gripper. You can change the size of the step by moving the red slider in the field display or by pressing a key from 1 to 9. After you finish with the first fiber type "q" or press the "Enabled" green button. At this moment the "Enabled" button will go red, the gripper will move to the second small fiber and the "Enabled" button will go green again. Repeat the process above until you finish all the small fibers.
After you finish logging the concentricities raw data you can use the "concenctio" program to generate a new concentricities file. [1] This program lives in /home/hydra/src/concenct. First you'll have to copy the old concentricities files to that directory plus the log file(s) containing the raw data. The old concentricities file will be used as a template to produce the new file. If the name of your current concentricities file is different than "concentricities" you will have to rename it as "concentricities". Then run the program to generate the new concentricities file. The name of the new file will be "nconcentricities". For example, to generate a new concentricities file from the current "concentricities" file plus the "hydra.currentLog" log file you should do:
% cd /home/hydra/src/concenct
% cp /home/hydra/Hydra/data/inits/concentricities .
% cp /home/hydra/fiblogs/hydra.currentLog .
% concen
Enter the name of the log file: hydra.currentLog
Small Concentricities
Group 1 Average is X= 0.000000 Y= 0.000000 for 135 measurements
Another file to read? (y or n): n
ave x = 0.000000, ave y = 0.000000, sdev x = 0.000000, sdev y = 0.000000
ave r = 0.000000, sdev r = 0.000000
var = 0.000000
var = 0.000000
var = 0.000000
ave x = 0.000000, ave y = 0.000000, sdev x = 0.000000, sdev y = 0.000000
ave r = 0.000000, sdev r = 0.000000
var = 0.000000
var = 0.000000
var = 0.000000
.
.
.
ave x = 0.000000, ave y = 0.000000, sdev x = 0.000000, sdev y = 0.000000
ave r = 0.000000, sdev r = 0.000000
var = 0.000000
var = 0.000000
var = 0.000000
%
If the data is contained in more than one log file, then copy all the log files to the concenct directory and answer yes to the question "Another file to read? (y or n):". The program will prompt you for the name of the second file, third file, etc.
When you run the program, make sure you also enter the large fiber and FOPS concentricity measurements (either a new set or old data took previously) to make sure that the zero point correction is handled appropriately.
Besides the nconcentricities file there are four other output files: raw.dat, ave.dat, con.dat and plot.macro. "plot.macro" is a supermongo script to generate individual plots for the concentricity measurement for each fiber. It uses the "ave.dat" and "raw.dat" files. File "con.dat" is for diagnostic value in case something looks strange. For example, to print the concentricity measurement plots:
% sm (sm is available in solaris and sunos workstations)
Hello hydra, please give me a command
: macro read plot.macro
: device postlandfile plotset1.ps
: plotset1
: device postlandfile plotset2.ps
: plotset2
: device postlandfile plotset3.ps
: plotset3
: device postlandfile plotset4.ps
: plotset4
: device postlandfile plotset5.ps
: plotset5
: device postlandfile plotset6.ps
: plotset6
Remember to place a comment in the header of the new concentricities file and to increment its version number. Then copy the new concentricities files to its final location at the Hydra PC and test at the telescope by configuring a standard field.
Last Modified: April 27, 2000
Before you can download code, you need to have the appropriate hardware and software configurations. Various switches and software items need to be configured. These are discussed below for both the host machine and the Galil Box.
a) Host Machine
The easiest way to download the code is to use the "Editor Tool" provided with the Visual Basic Tool. In order to use this software you will need your host machine to be running under Windows95 and the Visual Basic Tool installed.
b) Galil Box
While a comprehensive discussion of the Galil DMC-1500 is ledt to the Galil User Manual, a few pointers are given below.
In order to be able to do anything with the box, you have to be able to talk to it. To that end, a cable and the correct switch settings are required. No other hardware is necessary to talk to the box. Notice the little set of dip switches next to the serial ports on the black box. Set them as follows:
| switch # | Name | Position |
| 1 | MRST | OFF |
| 2 | 1200 | OFF |
| 3 | 9600 | OFF |
| 4 | 19.2k | OFF |
| 5 | HSHK | OFF |
This will set up things up for a baudrate of 9600 with no hardware handshake.
Now that you have the right hardware and software configuration you can start the Visual Basic Tool and get ready to download the code.
Start the Visual Basic Tool application and press the button labeled "Source Edit". The Editor Tool window will appear. Use the file directory window to browse for your makefile file. The Editor Tool use this file to load the files containing the code you are planning to download. This is a simple text file with a newline separated list of file names. If you are downloading the Galil Embedded Code [2] that file is "makefile.mak". It includes seven files: buttmove.g, coldstrt.g, errors.g, gripops.g, main.g, periphio.g and xyops.g.
Choose the makefile by clicking on it. The contents of the file will be showed in the "Make File Contents" entry. Here the Editor Tool gives you the opportunity to check the code in the files you just loaded before you attempt to make a download. Remember that each time you make a download the current code is lost. Click a file name in the "Make File Contents" box and the source code and declared variables will be showed.
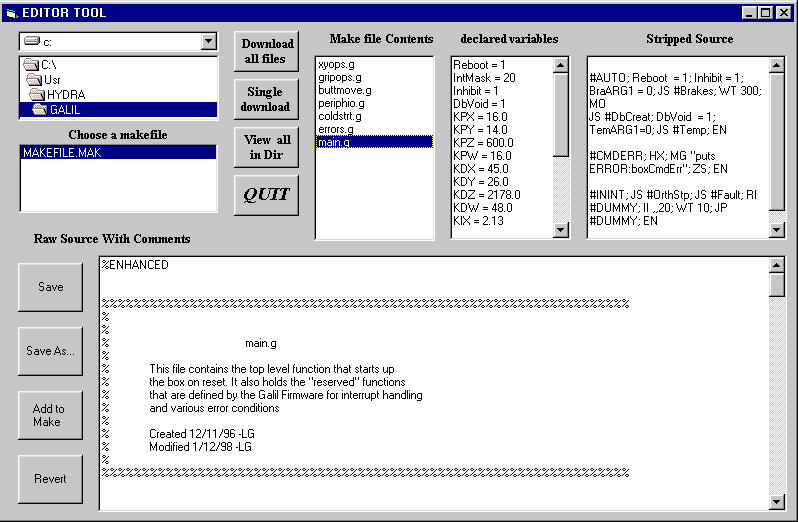
The final step is to download the code into the Galil. Press the button labeled "Download all files" to download the code. The download will take approximately 25 seconds. If everything went O.K. then a dialog box should pop up saying "Download OK".
Last Modified: April 27, 2000
Software binaries are available to run under Linux and Solaris. The latest versions will be found in hydrasimLinux [3] for Linux and in hydrasimSolaris [4] for Solaris. Both versions can also be found in the Tucson mirror of the CTIO ftp site [5] (updated nightly). Current version of the software is 4.0.2.
You will probably also want to download the the documentation file hydraassign.ps [6]. Thanks to Phil Massey for providing the documentation and software for the Hydra-CTIO version of hydraassign [7].
First, be sure you are running the c-shell or one of its derivatives (e.g. tsch). If you are running the bash shell, you can change to the c-shell using the command chsh -s /bin/csh Put the tar file in your home directory, change to your home directory and then
gunzip hydrasimLinux.tar.gz
tar -xf hydrasimLinux.tar
uncompress hydrasimSolaris.tar.Z
tar -xf hydrasimSolaris.tar
Now change to subdirectory "/Hydra/bin" and execute "~/install.org". This is a script that will create three sub-directories (fiblogs, fields & stdfields) in your home directory. Copy the file "~/Hydra/bin/sample.fiborg" to .fiborg in your home directory as well. Then include in the "set path" command of either your ".login" or ".cshrc" file the following path:
~/Hydra/bin
Also, add the folowing line to your .login or .cshrc file to source the .fiborg and set the environment:
source ~/.fiborg
Log out and back in to allow the new path to be activated or type "source ~/.login" (or source ~/.cshrc or whatever is your particular rc file). If you chose not to decompress and de-tar the downloaded file in your home directory change variable FIBHOME in file ".fiborg" to reflect the new location of the software as well as the bin path for the "set path" command.
Two windows will appear. One is a simple command line window (CLI/Script Tool window) and the other is a window showing a graphical representation of the fibers and the focal plate. It also includes various information about the current star field and status. The terminal window used to launch the program will be used as a console.
Last modified: October 17, 2001
Links
[1] http://www.ctio.noao.edu/noao/content/init-status-script-and-coord-files#concentricities
[2] http://www.ctio.noao.edu/noao/content/embedded-galil-software-files
[3] ftp://ftp.ctio.noao.edu/ctio/hydra/hydrasimLinux.tar.gz
[4] ftp://ftp.ctio.noao.edu/ctio/hydra/hydrasimSolaris.tar.Z
[5] ftp://ftp.noao.edu/ctio/hydra
[6] http://www.ctio.noao.edu/noao/sites/default/files/instruments/spectrographs/hydraassign.ps
[7] http://www.ctio.noao.edu/noao/content/hydraassign